What is the Role of Molecular Diagnostics in Infections Diagnosis in the Immunocompromised Host?
Molecular diagnostics, often based on PCR, are becoming more
common and widespread. The labor and material burden of traditional
culture-based diagnostics limits their use, while the emergence of
multi-directional panels that require little operator training explains the
technology's popularity.
Kits for the detection of viral, bacterial and fungal
pathogens were used in respiratory, gastrointestinal, blood and cerebrospinal
fluid samples, followed by drug susceptibility testing.
Several factors must be considered when considering molecular
diagnostics in immunocompromised hosts.
First, there is often a lack of extensive literature
assessing potential differences in sensitivity and specificity for
entrepreneurial entrepreneurship.
Second, immunocompromised hosts may have competing diagnostic
considerations in the stroma (eg, GVHD in blood transplantation), making the
issue of specificity critical.
Third, susceptibility testing often remains culture-derived,
limiting diagnosis to error-prone molecular tests until such tests incorporate
well-defined methods of resistance determinants.
Finally, lack of specificity can be an important drawback in
assessing positive rhinovirus or enterovirus because a particular virus or
strain may be relevant in different contexts (enterovirus D68 in blood or
transplant patients in recent outbreaks) Central nervous system disease in
patients with severe respiratory disease or coxsackievirus rituximab-induced
humoral immunity loss).
However, these sensitive tests improve our understanding of
the range of pathogens whose role in the disease process needs to be
understood. Furthermore, the sensitivity of molecular diagnostics for rapid
detection of pathogens to prevent nosocomial transmission is a major advance.
This is evidenced by a newly discovered pathogen (new smallpox virus associated
with equine exposure) or a previously undiagnosed pathogen (human
metapneumovirus).
How these susceptibility tests can help?
These susceptibility tests can help identify possible modes
of transmission and understand persistent infection with pathogens such as
influenza virus, norovirus, or varicella virus (vaccine strains), which may be
atypical and long-lasting in some patients.
Finally, new pathogens such as astrovirus VA1/HMO-C, which
have been reported to cause encephalitis, are now being identified using deep
sequencing, and the general application of this technology remains to be
determined.
Molecular detection of biologicals in this field has traditionally
been based on the use of quantitative real-time PCR (qPCR), which now includes
commercial instruments that can be used in the laboratory or in the field.
Bringing this technology into the latest applications
requires innovation to reduce size, weight and power requirements.
Rugged portable instruments, efficient power supplies,
lyophilized reagents, data communication, and standard operating procedures for
minimally trained users are examples of limitations that have been overcome to
generate qPCR-based data when needed.
Despite the high specificity and sensitivity of qPCR, these
assays require a priori sequence-based knowledge of the pathogen to design and
manufacture specific targeted assays using primers and probes. However, in many
cases the pathogen may be unknown and identification of the pathogen must be
based on the use of non-targeted screening methods.
By extracting, preparing, and sequencing all the genomic
material in a given sample at once (called metagenomics), a less biased picture
of the sample's biological entities can be established.
Deployment of a laboratory capability outside the clinical
laboratory walls is the necessary response to diseases that readily cross
international borders.
Applying metagenomic approaches in this field requires the
development and optimization of simple sample preparation, sequencing, and
bioinformatics workflows, reminiscent of the challenges faced in the development
of field-directed qPCR 15 years ago.
Due to the complexity and variability of biomaterials,
detection, identification and characterization of biomaterials, especially in
the field, is an ongoing challenge.
Traditionally, the most sensitive methods required the use of
culture-based methods that allowed identification of pathogens by demonstration
of viability. Culture-based methods require special equipment and laboratory
personnel, while early PCR is based on prior knowledge of potential pathogens.
In addition, culture-based pathogen detection requires a long
turnaround time (TAT), ranging from a few days to several months depending on
the organism. As a result, several culture-independent diagnostic tests
(CIDTs), especially those using nucleic acid amplification and next-generation
sequencing (NGS), have been developed to rapidly identify pathogens and are
more suitable for use in the field.
By molecularly classifying samples on demand (PON) and convenient transport of samples to a fixed laboratory environment, the TAT of pathogen detection is significantly reduced.
Many organizations, including
military defense, intergovernmental international, public and veterinary health
and law enforcement agencies, deploy field laboratories in a variety of
applications such as weapons of mass destruction (WMD) detection, infectious
disease outbreaks, and treaty validation.
As targeted detection limits pathogen detection to the number of targets included in the detection panel, further efforts are needed to fully develop unbiased sample analysis in this area.
These efforts should focus on the development of robust laboratory instruments, particularly size, weight, and power (SWaP), reagents that do not require refrigeration, and simple standard operating procedures.
What the most common technology in Molecular Diagnostics is based on?
The most common leading technology is based on the use of PCR, which was discovered by the late Curry Mullis in 1985 and revolutionized molecular biology. This discovery earned Mullis the Nobel Prize in Chemistry in 1993, and is designed to enable real-time analysis and quantification of target nucleic acids using real-time, or qPCR, instruments and the appropriate chemicals.
Real-time detection of target nucleic acids relies on the use of fluorescent, intercalating dyes or dual-labeled probes containing fluorophores and quenchers, such as TaqMan hydrolysis probes or singly labeled fluorescence resonance energy transfer (FRET) hybridization probes.
The most commonly used method is TaqMan, which uses Taq polymerase to amplify the target DNA and hydrolyze the double-labeled probes, thereby increasing the fluorescence detected by the instrument.
Alternatively, the use of FRET probes or intercalation dyes allows detection of target DNA and melting curve analysis (sample fluorescence vs. temperature), where post-PCR amplification produces melting peaks (first negative derivative of sample fluorescence vs. temperature) on a single nucleotide basis. sequences.
The availability of targeted qPCR assays, especially those developed and validated for commercial use, adds to the advantages of this technology, as suspect target reagents can be detected within hours with minimal hands-on time.
Real-time thermal cyclers are designed to be able to analyze up to 384 samples simultaneously in one run. In addition, the use of multiplex analysis allows the testing of multiple targets and the inclusion of internal controls in a single sample.
Alternatively, in-situ qPCR instruments are smaller, lightweight, robust, lower-throughput instruments that allow for field analysis of samples.
Since Watson and Crick identified the three-dimensional structure of DNA, much research and progress has been made in developing methods to read the genetic code of DNA molecules.
The advent of Fred Sanger's chain termination technique has revolutionized the ability to read DNA sequences. This pioneering Sanger sequencing method established first-generation DNA sequencing as the gold standard method for DNA analysis.
Second-generation sequencing, commonly referred to as NGS, has been instrumental in the introduction of massively parallel sequencing, providing higher sequencing throughput of millions of reads per second.
Currently, third-generation DNA sequencing, which is in its infancy, focuses on sequencing single DNA molecules.
Long-read sequencing technologies are now available to perform this innovative sequencing approach.
Advances in NGS protocols and instrumentation have made sample analysis faster and cheaper.
The use of NGS-based methods provides the advantage of unbiased metagenomic analysis of samples without prior knowledge of potential pathogens.
NGS-based sample analysis facilitates detection of multiple microbial infections, detection of antimicrobial resistance, monitoring and identification of new pathogens in a single assay. In addition, this approach allows identification of the pathogen in situations where other targeting methods may fail.
Despite these advantages, these methods have not yet been widely used in the field due to the need for refrigeration, large instruments, and trained bioinformaticians. In recent years, commercially available handheld devices capable of on-site sequencing have been able to detect pathogens in less than 6 h.
Adherence to operational limitations in this area, such as remote feeding, elimination of freezers, and incorporation of a simple, intuitive product design that allows minimally trained users to perform tests and report results. However, further development of sample preparation and data analysis is required to enable sequencing for routine sample analysis.
As qPCR and NGS technologies continue to evolve, NGS is generally less practical in the field than current qPCR products and their operating concepts.
Currently, qPCR still maintains higher analytical sensitivity, specificity, reproducibility, and ease of use than NGS. In addition, qPCR offers advantages in terms of usable result processing time and lower cost per sample.
However, NGS retains the advantages of detection power and scalability in the number of markers that can be analyzed simultaneously.
Considerable progress has
been made in addressing SWaP, as well as the portability and robustness of
performing qPCR and NGS in field distribution settings.
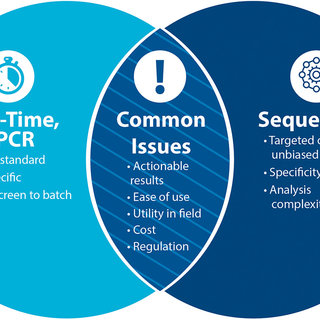 |
| Comparing qPCR and sequencing-based molecular methods |
Products and related systems developed for cross-border applications in this field, including qPCR and NGS-based technologies, from companies and artists who can be called "first movers and fast followers" derived from industrial economic models suitable for industrial economic models.
The first mover is often considered to be the ringer, the trailblazer, and the first to market, acting as the tip of the spear. They have successfully applied new technologies through proprietary research and development concepts, accelerated product development, especially early end-user champions, and strategic sales, business marketing and successful market entry.
Simon Sinek created the Law of Diffusion, a theory of how, why, and how quickly new ideas and technologies spread, with early movers and fast followers at the forefront of an ecosystem.
Fast Followers, while below market, have the first skills and are still competitive and nimble. Previously, the military and defense focused on the capabilities and requirements to detect biological agents in the field, and the 2001 US anthrax attack expanded this, leading to a broader US government effort to establish a biodefense enterprise and associated security application.
A balance of performance discriminators including ease of use, portability, robustness, speed, sensitivity, specificity, reproducibility, detection power, and throughput is required for qPCR and NGS products to be successful in this field.
Costs are also important for consumables such as testing, life cycle, instrument maintenance/warranty and operator training.
(i) qPCR. One of the pioneers in molecular testing of biologicals in this field is BioFire, formerly Idaho Technology, Inc. BioFire is optimizing an existing laboratory qPCR platform called R.A.P.I.D. for cross-border applications in this field.
From the mid-1990s to the early 2000s, this optimization focused on robust instrument manufacturing and development of lyophilized reagents and automated software analysis to aid on-site logistics and improve ease of use.
BioFire also implements three different military field PCR systems (RAZOR, JBAIDS and FilmArray) for FDA-licensed biometrics, including the Clinical Laboratory Improvement Amendments (CLIA) exemption system. In the mid-2000s, they collaborated with the US Special Operations Command to develop the RAZOR man-portable field system. They incorporate features from previous generations of field-ready qPCR systems (robust, lyophilized reagents and automated assays), reduce system footprint, and increase battery power.
The most important achievement they have achieved is its ease of use, which eliminates pipetting and reduces the number of steps required to obtain results.
Over the past decade, BioFire has further integrated its systems with sample preparation, increasing multiplexing capacity while increasing ease of use and reducing the steps required to obtain results.
Future technologies include "Extreme PCR," a phrase coined by its inventor Carl Witwer (co-founder of Idaho Tech and BioFire), which can reduce qPCR run times from tens of minutes to tens of seconds of technology. While the current time-to-result (TTR) of 45 minutes for multiplex CLIA tests is acceptable, it does not fully meet the needs of the point-of-care market.
(ii) Sequence. Sort Space has seen several iterations of Forerunners and Fast Followers, many of which have become early casualties and quick burials. The most striking and notable examples of the latter have been mainly of the second generation order. Pyrosequencing (eg 454), semiconductor sequencing (eg ion torrent) and sequencing by ligation (eg SOLiD) offer unique advantages with their technologies and short time to market. However, their advantages cannot surpass the quality and reliability of first-generation chain-end sequencing (which some argue is still the gold standard), or compete with third-generation throughput, read length, and low read cost. basic platforms such as sequencing-by-synthesis (ie, Illumina), single-molecule sequencing (ie, PacBio), or nanopore sequencing (ie, Oxford Nanopore Technologies [ONT]). But even among the third-generation products that have dominated the market for the past 5 years, important differentiators are emerging that indicate where the sequencing market is headed and how sequencing data will be used in the next 5-10 years.
Among the leading third-generation platforms described above, ONTs can be considered the forerunners of on-site sequencing or sequencing outside of traditional brick-and-mortar laboratories at or near the point of sample collection.
It is the first and so far the only sequencing platform that can reasonably be considered portable. The paradigm shift associated with portable sequencing focuses on the ability to generate sequence-based information on demand, rather than traditional hospitals, advanced clinical facilities, or research laboratories. In other words, the positive health outcomes, biosphere stewardship, and natural product discovery benefits associated with genome sequencing will no longer be limited to those fortunate enough to live near more traditional research facilities.
Is there any option to pass the lab to the sample instead of the other way around
There is now an option to pass the lab to the sample instead of the other way around. But to fully understand the small size of the new sequencing hardware, there are still development challenges: you still need to buy your own lab.
Front-end sample preparation (ie, nucleic acid extraction, target enrichment, library preparation, etc.) and back-end data analysis (ie, bioinformatics, taxonomic assignment, sequence assembly, functional annotation, phylogeny, etc.) lag behind. portability and ease of use Sorting hardware.
A complete end-to-end portable sequencing process takes much more than the hardware itself.
To effectively support and execute the entire process:
(i) powerful portable power supplies are required to support
(ii) the massive computing power required for base calling and sequence data analysis
(iii) flow cells and refrigerated solutions for sequencing reagents
(iv) ) ergonomic lab bench to perform manual workflows and access various pipettes, pipette tips and other consumables,
(v) biosafety and biohazard storage/disposal facilities and
vi) accessory hardware/equipment for library preparation (e.g. thermal cyclers, microcentrifuge), etc.).
ONT has developed additional hardware to address some of these issues, namely the Voltrax device (an automatic library preparation device) and the MinIT (a GPU-based computing device that integrates the MinION operating system and fast base calls). Recently, ONT developed an on-site sequencing kit consisting of lyophilized reagents that do not require refrigeration.
Third-party developers have begun to innovate additional solutions, such as FPGA-mediated base calling to improve MinIT scalability, algorithm optimization for more efficient nanopore data analysis, and logistical organization. configuration of a one-person portable mobile laboratory structure like http://www.mriglobal.org/2018/11/08/mercury-labs/.
Rapidly taking advantage of the first generation of ONTs, such as integrated device manufacturers such as Ontera, manufacturers of nanopore-based molecular detection platforms in a portable format.
Although not a definitive sequencing technology, the platform offers a new method to detect molecular signatures from a variety of targets, including DNA, RNA and proteins, in a situation where it is needed.
Streamlined push-button bioinformatics software with a graphical user interface (GUI) and standardized reporting nomenclature and automation of wet lab processing steps (e.g. DNA/RNA extraction from various sample matrices, molecular enrichment, sequencing) continue to hold significant potential for innovation in library preparation, etc. .). This innovation is necessary to transfer this technology to routine biomonitoring and clinical settings related to regulatory frameworks.
The regulatory framework itself will need to evolve to accommodate new markets, such as sequencing-based on-site diagnostic testing, point-of-sampling for food safety and agricultural surveillance testing, supply chain certification testing at the point of entry, and forensic personal identification. to name a few.
The ethical politics surrounding human DNA testing have not kept pace with practical sequencing on the ground. Recent DNA testing on the US-Mexico border, as well as consumer DNA testing in general, has highlighted the need for norms around consent, acceptable provenance, and the legal use of personal genomic data.
Given the massive diversity of samples, genome-based diseases, the functional utility of genomic information in biomonitoring, and the lack of regulatory and political consensus on the generation and use of genomic information in new settings, a strong new market is expected to address these concepts so as to develop rapidly in the future 10 years of growth.
A retrospective example of this field: The problem of weaponized anthrax. After the first and second Gulf wars, the United Nations established two special committees: the United Nations Special Committee (UNSCOM) and the United Nations Monitoring, Inspection and Verification Committee (UNMOVIC) to inspect, verify, destroy and monitor Iraq's weapons of mass destruction. (WMD).
Iraq produced several biological agents and weaponized them by loading certain types of munitions, such as R400 bombs.
The inspectors approach the task in a forensic-like manner, interpreting and verifying Iraqi declarations of production, weapons and unilateral destruction. Although UNSCOM's work often depends on the decisions of the UN Security Council and related diplomatic and political factors, their experts in the field are very quick to take advantage of the introduction of new technologies.
In 2002, UNMOVIC deployed a mobile laboratory with chemical and biological analysis capabilities. When it comes to bioanalysis, labs rely heavily on qPCR (Idaho Technology), immunoassay technology, and other technologies funded by various governments.
In 2002, UNMOVIC received permission to excavate the site of what was reported to be the destruction of an R400 bomb filled with anthrax.
Inspectors found eight intact bombs filled with liquid suspected to contain botulinum, aflatoxin or anthrax, which were said to have been used to fill the R400 bombs. Confirmation of R400 bomb content is done through primary, secondary and tertiary processes.
The initial validation process begins with safe drilling to determine if the R400 is full of chemical warfare agents, then samples are taken to a field laboratory and tested for chemical warfare agents and enzyme-linked immunosorbent assay (ELISA) and PCR methods to identify biological agents.
In this case, the potassium permanganate used by the Iraqis to disable bombs loaded with biological agents challenged the PCR analysis as a chemical interferent.
In addition to chemical interference, early
challenges included performing DNA extractions, which were limited by the
quality of the kits currently in use. Figure 4
Figure 4 R400 bomb excavated from the Al Azizia site. On the
left side of the image is a prototype drilling device called MONICA. (Photo by
Kay Mereish.)
Secondary and tertiary validation processes are carried out in reference laboratories following standard chain of custody procedures, while the main process is carried out on site by UNMOVIC inspectors.
Two international reference laboratories tested the samples for chemical and biological warfare agents, and the results confirmed the presence of potassium permanganate and Bacillus anthracis DNA.
The results of the reference laboratory analyzes have been communicated to the Security Council.
The genome of Bacillus anthracis was identified using multilocus variable number tandem repeat (ML-VNTR) analysis published in 2000.
In retrospect, the UNMOVIC field laboratory deployed to test biological warfare agents was the first of its kind to successfully conduct analyzes in a field boundary environment, and samples were safely transported to reference laboratories for validation.
In 2003, UNMOVIC proposed the use of qPCR analysis at the Al Hakam facility, a bulk pharmaceutical disposal site in Iraq. Although the inspectors were unable to convince the UN to approve their proposed field use protocol, their field laboratory results compared to those of a reference laboratory could be used as a model to support ongoing negotiations to create a validation protocol for an ad hoc biological weapons convention. (BWC) Panel of Experts.
There are currently no further negotiations or plans to enter into a BWC-type inspection agreement. Other experience in this field. In addition to examples of tract validation using qPCR in the field and confirmatory sequencing in a reference lab, our panelists describe their experiences as fast followers in the field. Examples of military (clinical diagnostic) and wildlife health applications are described. Their experience using primarily qPCR platforms reflects the need to quickly identify biological needs and targets and to use established products such as assays to address PON. i) 2014-2016 Ebola virus outbreak. When U.S. troops deployed to West Africa in 2014 to respond to the Ebola outbreak, they faced enormous challenges managing a workload that would eventually total more than 32,000 samples between 2014 and 2016.
The EZ1 Ebola Zaire test, developed by USAMRIID's Diagnostic Systems Division, is being used by the US Army, NIAID, and a local national laboratory team working at the Liberia Biomedical Research Institute to address this need. Thanks to some forward thinkers, the FDA has designated the EZ1 test as a "Pre-EUA" test for the BioFire JBAIDS, Roche LightCycler, and ABI 7500 Fast DX systems. It was quickly updated and then received FDA Emergency Use Authorization (EUA) in August 2014, making it the first EUA Ebola test kit available to US personnel in affected areas. The ability to develop content on an open platform is critical to the success of EZ1 Analytics. This demonstrates the clear benefits of a more open and practical platform, and within limited resources, this development philosophy complements other efforts to completely eliminate in vitro diagnostic (IVD) devices. More than a decade ago, the vision of the Department of Defense Diagnostics Development Command, combined with a large number of personnel willing to deploy to the region and the commitment of local laboratory personnel, successfully addressed the 2014-2016 challenges of the 2018 Ebola outbreak.
(ii) US Department of Defense regulations define five roles or levels of medical care that a patient can receive in an emergency:
- Role 1 (unit level),
- Role 2 (emergency trauma care)
- Role 3 (medical facility)
- Role 4 (hospital). . a level)
Another example of military and defense experience involves a deployment to Afghanistan based at a level 3 medical facility in Afghanistan.
Blood banking and blood chemistry, which can be measured using hand-held devices such as the Abbott Piccolo and i-STAT, are a priority for Role 3 medical facilities. BioFire JBAIDS allows us to create an on-site diagnostic system.
The first challenge was to find space in a Conex-type mobile
laboratory to interpret microbiology and molecular biology, which required
clean and dirty areas.
The next challenge in the implementation is to get familiar
with the technology and related operating concepts, that is, to organize and
quickly install all the necessary components and materials. In demanding and
unforgiving environments, simple operational tasks such as computer password
management and day-to-day communication problems become time-consuming. Two
JBAIDS tools are used: one is functional and one is for spare parts. Overall,
the system is very compact, but materials such as extraction kits and various
boxes of consumables are still larger than what we use in biochemistry today.
Cold chain storage is unreliable and almost non-existent. Despite the
operational challenges, the technology helped us achieve results relatively
quickly, but it required technical expertise. (iii) Understanding of disease
transmission in wild animals. Wildlife health testing in remote areas was a
third experience mentioned by one of the discussants. An example of
Campylobacter testing in wild mountain gorillas demonstrates that qPCR
technology is useful and practical for early detection of pathogens and
containment of potential outbreaks (37). As mentioned above, the ability to
perform qPCR and sequencing in the field reduces result TAT, which also
benefits wildlife by rapidly informing treatment, possibly even while the
animal is still anaesthetized. Similar challenges in performing qPCR in the
field are also noted, such as instrument power and lack of cold chain storage.
Early results are valuable when animal samples can be reliably obtained and
analyzed in situ using qPCR, often reducing or eliminating complex import and
export requirements for animal samples that may be subject to various
international trade regulations such as CITES. A panelist who studied during
his Ph.D. pointed out an important challenge. student, still challenging it as
a Ph.D. The professor is the cost of lab equipment, whether it's qPCR or sequencing
equipment. Fees remain a budget barrier for most academics studying wildlife.
He emphasized the importance of borrowing and demonstrating the procedures,
allowing investigators to borrow otherwise expensive tools to expand the use of
these techniques, improve diagnostics in wildlife, and understand how pathogens
interact with their hosts. Panelists also reminded attendees that these methods
lend themselves well to One Health's approach to identifying shared health
issues between animals, humans, plants and the wider environment, better
discovering and investigating shared infectious disease pathogens and their
natural ecology. Workshop opening
Our group has a two-fold goal: to promote discussion and increase understanding of
(a) the technical aspects of qPCR and sequencing, which share similar direct and targeted approaches, NGS offers greater discovery opportunities, and
(b) the operational aspects of performing these activities. methods in situ, they are parallel, and qPCR is probably more mature.
From our discussions, including questions from the audience, our group identified the following points related to utility and future challenges.
Current qPCR and sequencing methods are complementary and interdependent. Although orthogonal methods such as immunoassays have value, qPCR systems designed for field use are the most powerful because of significant investment in optimized chemistry and instrumentation (SWaP).
qPCR and sequencing technologies are related and interconnected, for example, strains must be sequenced to develop new and improved qPCR assays.
In addition, the robust sequencing of qPCR and PON overcomes the hurdles of sending samples to countries with licensing issues and sensitivity.
The advent of funding for biodefense has attracted scientists, particularly in the life sciences, to entrepreneurship.
Related industries, such as environmental testing, also address biodefense needs. As lower costs allow end-users to make decisions, manufacturers still need ongoing funding opportunities and incentives to continue their R&D in other reactive markets to overcome the "valley of death" for their products. At the other end of the spectrum, one panelist noted the importance of prioritizing underrepresented research topics such as environmental and wildlife biology. Funding opportunities are few and often classified as larger clinical opportunities. Maintaining valid qPCR assays is an ongoing challenge.
The effect of erosion of function is that the established qPCR assay reduces sensitivity and specificity when testing against new strains.
The quality of qPCR assays depends on continuous testing and evaluation of new isolates and/or known strains that have mutated in nature.
For retrospective laboratories with high-throughput sequencing capabilities, this requires time and resources.
In addition to the sample suppression mentioned in the UNMOVIC example, sample size imposes test limitations, so non-destructive analytical methods are also required to accommodate the various biological and chemical methods.
As an example from a clinical diagnostics perspective, BioFire Defense is developing a panel for tropical diseases that can detect approximately 20 pathogens simultaneously, requiring significant time for testing, validation, institutional review boards (IRBs) and clinical trials worldwide.
It is long and the intended use of the diagnostic analysis is carefully defined. As outbreaks develop, sequencing data is reviewed to see if the test is effective in detecting pathogens. Although this is part of product maintenance, there is often a delay between an outbreak and the availability of sequence data or samples. This delay is compounded by geopolitical pressures on sample and information sharing, as well as concerns about reportable diseases.
qPCR and sequencing platforms become more accessible, people will get creative with testing different sites.
When unexpected diseases are discovered, there is often no standard policy. Then an interesting problem arose where customers often asked to "hide" target results in our dashboards if they didn't have a policy to handle positive results.
Finally, the jury strongly agrees that greater scientific transparency is mutually beneficial. Although it is more convenient to share sequence data via shared transfer, the well-intentioned need for better sharing of isolated materials remains important for continued scientific transparency.
In general, it starts with cooperation and building
trust in a coordinated bilateral way. Examples include partners conducting
research while actively engaging with partner governments, giving them input
and ownership. Liberia after 2014-2015 The 2010 Ebola outbreak is an example.
We must all collectively attempt for regular scientific transparency.
Author's Bio
Education: MBBS, MD
Occupation: Medical Doctor
Specialization: Community Medicine, General Surgery, Natural Treatment
Experience: 18 Years as a Medical Practitioner
Share this in Public Information Interest>>



Comments
Post a Comment